|
Tool
|
Brief Description, Possible Uses, and URL
|
|
Charts
|
|
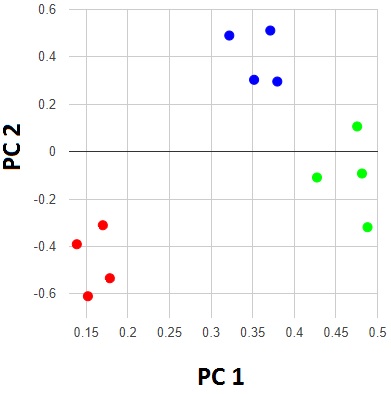
Google Charts, uvCharts, Chartist.js
|
They offer very complete sets of chart types, and are very customizable.
|
|
ChartJS, Highcharts JS, Flot
|
Also very complete, but here grouped together because they all provide support for older web browsers, yet deliver high-quality graphics, interactivity, and animations in modern browsers.
|
|
Smoothie Charts
|
Especially suited for plotting streamed data in real time.
|
|
D3.js, n3-charts, Ember charts
|
D3.js has very broad applications, which allows charting in multiple formats not covered by the libraries mentioned above. Libraries like n3-charts and Ember charts make plotting through D3.js easier.
|
|
Mathematics and statistics
|
|
NumCalc.com
|
A very complete scientific calculator online.
|
|
LALOLib and Mlweb
|
Very complete libraries for mathematics in the browser, with tools for advanced matrix algebra, statistics, optimization, and machine learning. Can be used as libraries for web apps or through a complete environment (LALOLab) similar to that of stand-alone math programs, with its own plotting capabilities.
|
|
Numeric.js
|
Another powerful library for mathematics in the browser, also usable through an environment similar to that of stand-alone math programs.
|
|
mljs
|
Library for machine learning.
|
|
JavaScript LIBSVM
|
C++-to-WebAssembly port of the LIBSVM support vector machine library
|
|
ConvNetJS, synaptic.js, brain, mind, DN2A
|
Libraries for neural networks and deep learning, covering from simpler utilities like data and function approximation and regression, to self-organizing maps, image regression and object identification, deep learning, and linguistics. Most can be trained and used online, or trained offline and used online.
|
|
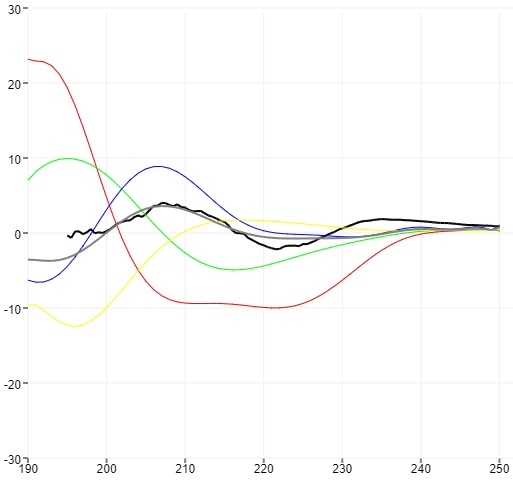
jsfft, FFT.js, FFT in many languages, DSP.js, timbre.js
|
Libraries for Fast Fourier Transforms; together cover direct and inverse transforms, real and complex convolutions, discrete transforms, and other functions (plus, some are actually full packages for signal processing).
|
|
Math.js, Sushi, numbers.js, jstat, rift, science, glMatrix
|
Other math libraries, some with unique features or tailored for specific applications (for example, rift is intended for games, glMatrix, and Sushi for efficient matrix calculations)
|
|
Strings
|
|
String.js
|
Functions that extend those of standard JavaScript strings.
|
|
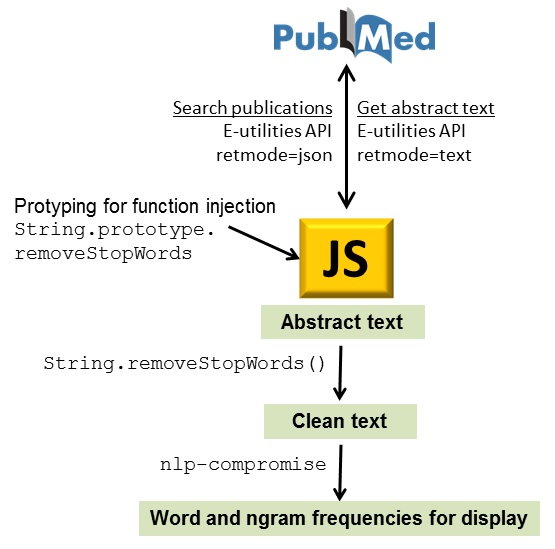
textmining and text-miner
|
Two similar packages for text mining. Potentially useful for mining contents in large corpuses of scientific text; Figure 1B of this article was built using such techniques. Text-miner includes lists of stopwords in four languages.
|
|
RiTa.js and nlp-compromise
|
Two libraries for natural language processing.
|
|
User interfaces
|
|
WebGazer
|
Eye tracking library that uses common webcams to infer eye-gaze locations of the user on the web page in real time. Could be used to facilitate molecular visualization as shown in this proof-of-concept example.
|
|
Tangle
|
Library to create reactive documents, i.e., which interact with the user in a contextual manner and virtually immediate response.
|
|
MathJax
|
Library to display formatted maths in the browser.
|
|
Emulators and programming
|
|
Linux emulators by Bellard and Macke
|
Two complete emulators of a linux system, the second including internet connectivity and a graphical interface. Could be used for learning linux in a safe and simple environment for example at the beginning of a tutorial course that requires linux knowledge; also for executing shell commands and scripts as well as programs written for Perl, Python, or even compiling C code, etc. on non-linux computers without the need for complex installations, as exemplified with the NESmapper Perl script.
|
|
Online programming environments
|
The two linux emulators listed above include C compilers and allow running Perl, Python, etc. Moreover, there are JavaScript-based web applications specifically tailored for writing, compiling, and running programs. Some notable links: site for learning and running C at http://cs-education.github.io/sys/#VM ; a Perl interpreter at
https://gfx.github.io/perl.js/ ; see also Emscripten at http://emscripten.org/
|
|
Vi emulator
|
A working online version of the linux vi editor, could be used to quickly process text files in non-linux computers.
|
|
3D graphics, computer vision, and augmented and virtual reality
|
|
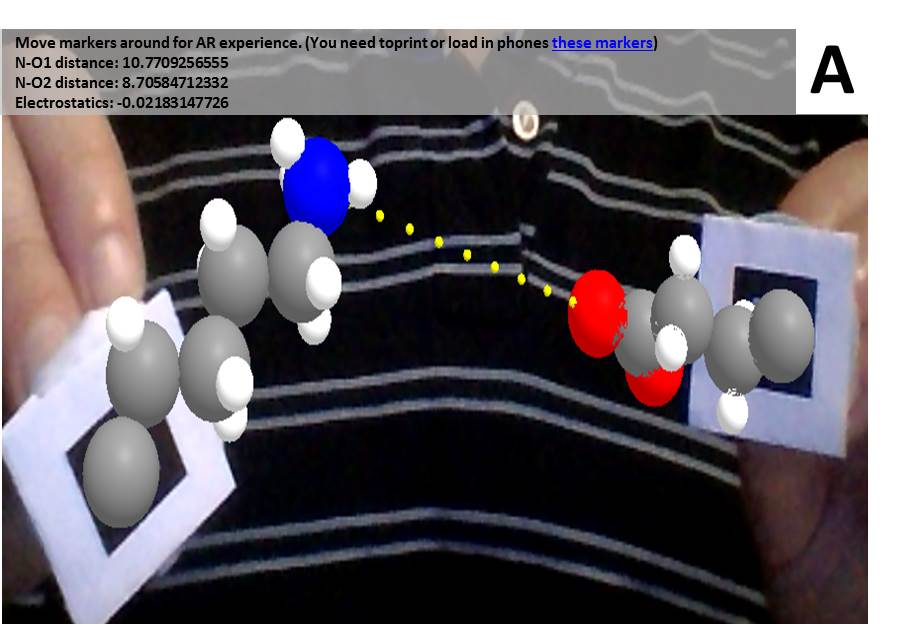
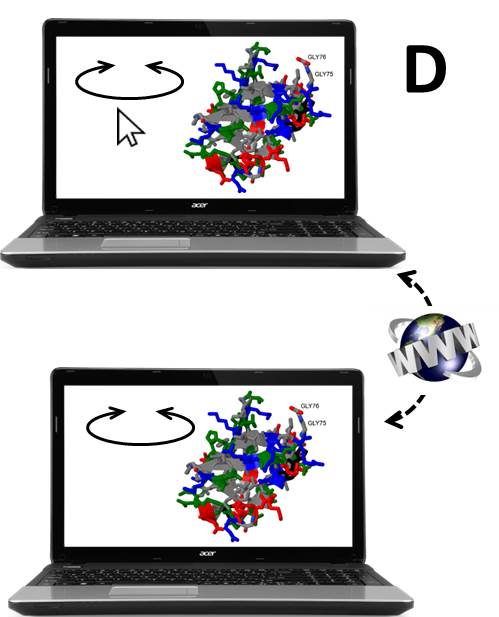
Three.js,
A-Frame
|
Three.js is probably the most used library for animated 3D graphics, using WebGL. Applications are in molecular visualization and in more generic data visualization, but beyond these uses, it includes multiple numerical algorithms that could be recycled for other purposes.
A-Frame is an entity component system framework for Three.js, that makes it much easier to use in virtual and augmented reality applications, the latter especially through AR.js.
|
|
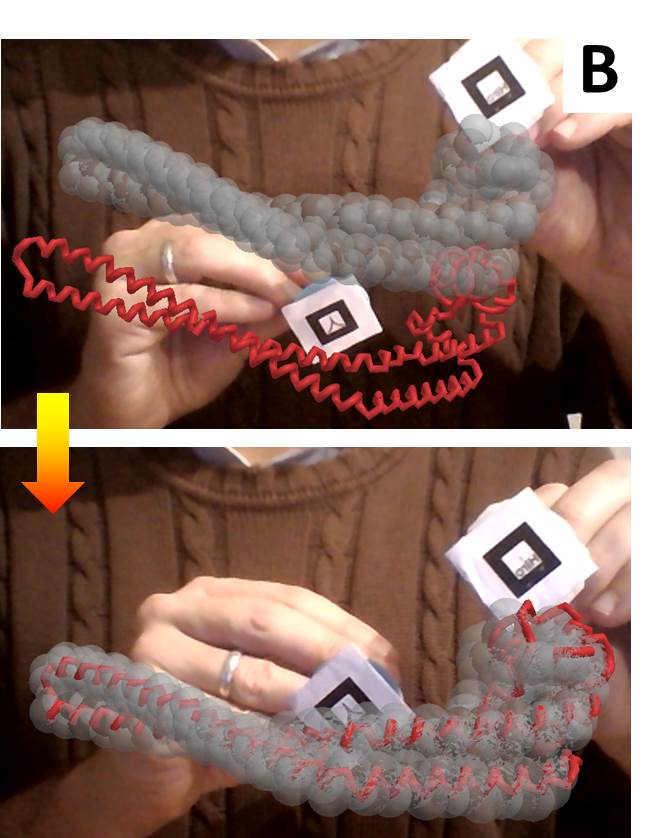
Jsfeat, tracking.js, js-aruco, AR.js, awe.js, and argon.js
|
Computer vision and image processing libraries which can identify and track objects or markers. Just like Three.js, they include numerical algorithms that could be used for other purposes. Put together with graphical libraries like Three.js leads directly to augmented reality web apps.
|
|
image-js
|
Efficient image processing and manipulation in JavaScript
|
|
Tesseract.js, Ocrad.js
|
Libraries for optical character recognition from images, useful for retrieving information from article figures.
|
|
Hardware and communication
|
|
WebRTC
|
Not a library but a collection of standards, protocols, and APIs for real-time server-less, plugin-free communication and data exchange directly between web browsers.
See also the example at https://apprtc.appspot.com/ which implements a web app for server-less video conferencing.
|
|
PeerJS
|
Browser-to-browser transfer of data, video and audio. Potential applications for concurrent visualization and collaboration.
|
|
HTML5 guitar tuner
|
A web app that guides guitar tuning; exemplifies how to read microphone data and analyze the signal through Fourier transforms to decompose the frequency spectrum.
|
|
annyang!
|
Speech recognition library to control web page with voice commands.
|
|
gyro.js
|
Simplified access to gyroscope and accelerometer information, expanding the information provided by the built-in Geolocation API (which only locates the user’s position based on network and/or GPS data but gives no orientation information).
|
|
GPS.js
|
Access to primary GPS data.
|
|
gpu.js, turbo.js,
|
Libraries to run calculations on Graphical Processing Units (video cards)
|